Details of the Drug
General Information of Drug (ID: DMFO843)
| Drug Name |
Fluspirilene
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Fluspirilen; Fluspirileno; Fluspirilenum; Imap; Redeptin; Spirodiflamine; R 6218; F-100; Fluspirileno [INN-Spanish]; Fluspirilenum [INN-Latin]; Imap (TN); Spiropiperidine analogue, 1; Fluspirilene (USAN/INN); Fluspirilene [USAN:INN:BAN]; McN-JR-6218; 1-Phenyl-4-oxo-8-(4,4-bis(4-fluorophenyl)butyl)-1,3,8-triazaspiro(4,5)decane; 8-(4,4-Bis(p-fluorophenyl)butyl)-1-phenyl-1,3,8-triazaspiro-(4.5)decan-4-one; 8-[4,4-bis (p-fluorophenyl)butyl]-1-phenyl-1,3,8-triazino[4,5]decan-4-one; 8-[4,4-bis(4-fluorophenyl)butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one; 8-[4,4-bis(4-fluorophenyl)butyl]-4-phenyl-2,4,8-triazaspiro[4.5]decan-1-one; 8-[4,4-bis(p-Fluorophenyl)butyl]-1-phenyl-1,3,8-triazino[4.5]decan-4-one
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Therapeutic Class |
Antipsychotic Agents
|
||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
||||||||||||||||||||||
| ATC Code | |||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
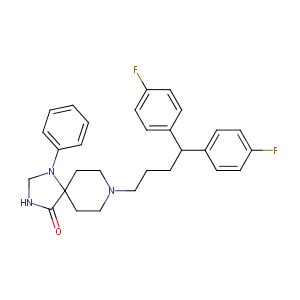 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 1 | Molecular Weight (mw) | 475.6 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 5.6 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 7 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 5 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | ||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | |||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Schizophrenia | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | 6A20 | |||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||||||||
References
